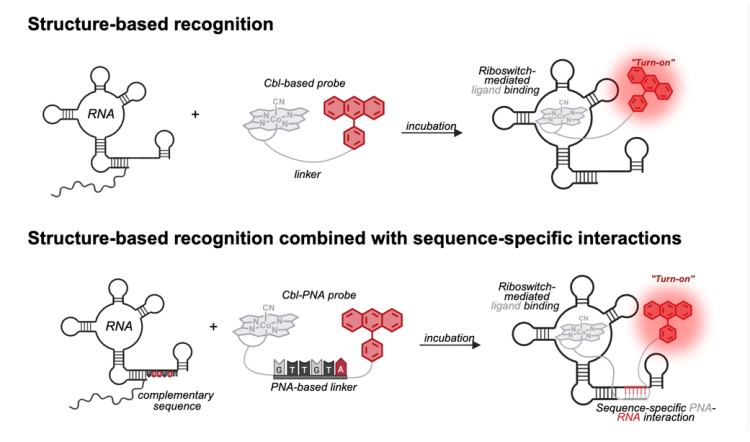
Developing RNA imaging tools and making tools out of RNA

RNA carries genetic information and performs diverse structural and catalytic roles in the cell. It can fold into defined three-dimensional shapes, allowing it to regulate gene expression, catalyze reactions, and interact with proteins and small molecules. It’s unique features made this molecule well-suited for developing genetically encoded probes and sensors because it can fold into diverse, highly structured architectures capable of binding specific small molecules, proteins, or ions. Its sequence-structure relationship allows precise tuning through rational design or in vitro evolution methods, and naturally occurring riboswitches provide proven templates that can be adapted or re-engineered to broaden the range of detectable targets. RNA can be expressed directly in living cells, enabling real-time sensing, and its modularity allows seamless integration of sensing domains with reporter elements to generate signals through fluorescence, conformational changes, or regulation of gene expression. Because it is compatible with standard molecular biology tools, RNA can be readily incorporated into plasmids, viral vectors, or genomes. Finally, RNA sensors can operate in contexts where proteins may misfold or be challenging to engineer, making them a versatile platform for next-generation genetically encoded probes.