Developing RNA imaging tools and making tools out of RNA

RNA Research Projects
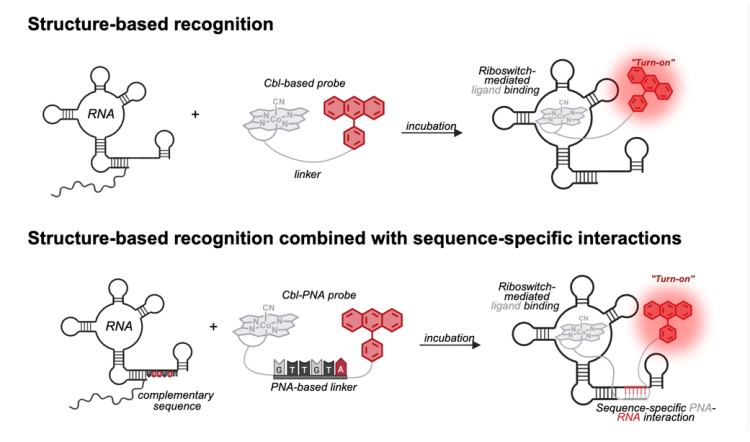
About three-fourths of our genome is transcribed into RNA, and these RNAs are involved in essentially every cellular process. Like us, RNA cannot do its job if it is not in the right place at the right time, so the transport of RNA to its proper place is critical for cellular function. Since the 1990s, scientists have figured out how to add fluorescent tags to RNA so that we can watch the RNA move around in the cell in real time under a fluorescence microscope which makes investigating complex research questions easier to pursue. Riboglow, a collaboration between the Palmer and Batey labs in the Department of Biochemistry at CU Boulder, is a new take on these RNA-imaging tools based on naturally occurring RNA sequences, called riboswitches, that fold robustly in the cellular environment and bind their ligands tightly and specifically. Like many other RNA-imaging systems, Riboglow is a two-part system made up of an RNA component (cobalamin riboswitch) and a fluorescent component cobalamin-based probe). The sequence of the RNA component, the aptamer, can be fused to the 3' end of the gene of interest, similar to the addition of a FLAG tag or GFP to a protein of interest. When transcribed, the final RNA includes the RNA of interest and the aptamer. The fluorescent component, or probe, contains the ligand for the riboswitch sequence that the aptamer is based on, a linker, and a fluorophore. Then, this aptamer can tightly and specifically bind the fluorescent probe, completing the fluorescent tag on the RNA of interest. We explored different types of linkers within our fluorescent probe and demonstrated that synthetic oligonucleotides (in this case, peptide nucleic acids) can function as an additional binding motif. This establishes a new route for improving RNA imaging tools and highlights the potential of short oligonucleotides conjugated to small molecules to enhance the affinity and specificity of RNA-targeting chemical probes.
A multicolor riboswitch-based platform... Unveiling the promise of peptide nucleic acids... Enhancement of RNA Imaging Platforms...
Fluorescent biosensors have transformed how we study dynamic biochemical processes, enabling direct visualization of events in live mammalian systems. However, most existing biosensors rely on fluorescent proteins, which suffer from limited color diversity, broad spectral overlap, and modest sensitivity-factors that restrict multiplexed imaging and accurate quantification. To address these challenges, our work aims to develop a modular RNA-based biosensor platform capable of expanding the available color palette, improving signal-to-noise performance, and providing new opportunities for studying targets with spatial and temporal precision. Nucleic-acid–based biosensors offer a strong path toward practical, genetically encodable sensing tools. RNA is especially useful because its structure can be evolved to recognize a chosen small molecule. In addition, naturally occurring riboswitches already form defined architectures that bind a wide range of molecules with high specificity. These riboswitches can be re-engineered to reshape their binding pockets, expanding the set of ligands they can detect.
Structural basis for ring-opening fluorescence by the RhoBAST RNA aptamer

