Home
Research Highlights
[video:https://www.youtube.com/watch?v=9tAIbpZ_uOw]
"Photoactivated antibiotics to treat intracellular infection of bacteria" on the use of Quatum dot based antibiotics to reduce intracellular infections in a bone-infection model.
Infrared-light activated Quatum dots kill MDR bacterial infection in a mouse model. The study shows that QD antibiotics are safe and effective in an abcess infection model.
Quantum dot antibiotics can treat biofilms of multi-drug resistant bacterial isolates of MRSA, Psuedomonas aeruginosa, and Escherichia coli.
Quantum dot antibiotic animal safety and efficacy study "Photoexcited quantum dots as efficacious and nontoxic antibiotics in an animal model" shows that specifically-engineered and light-activated Quantum dot antibiotics can treat infection of MDR bacteria in an in vivo model, and are safe and non-toxic.
Near-infrared-active quantum dots (QDs) have been designed to generate therapeutic superoxide radicals within pathogens. The combination of activation using a deep-tissue-penetrating light source, and selective mechanism of action, produces an effective treatment.
Facile and Accelerated Specific Thaerpeutic (FAST) platform is used to develop antisense molecules that target genes of interest in non-model or genetically intractable spcies, such as MDR clinical isolates of bacteria. This allows us to identify which genes are important for antibtioic tolerance and/or resistance. This process can identify new gene targets in MDR bacteria in less than 5 days.
Transcriptomic Response to Superoxide Generating Quantum Dots in Echerichia coli confirms mechanism of action of Quatum dot antibiotics. We identify genes demonstrate a consistent association with the DNA damage response and deactivation of iron–sulfur clusters.
Potentiating antibiotic efficacy via perturbation of non-essential gene expression shows that combining individually fitness-neutral perturbations can compound fitness impacts and sesitize bacteria to antibiotics.
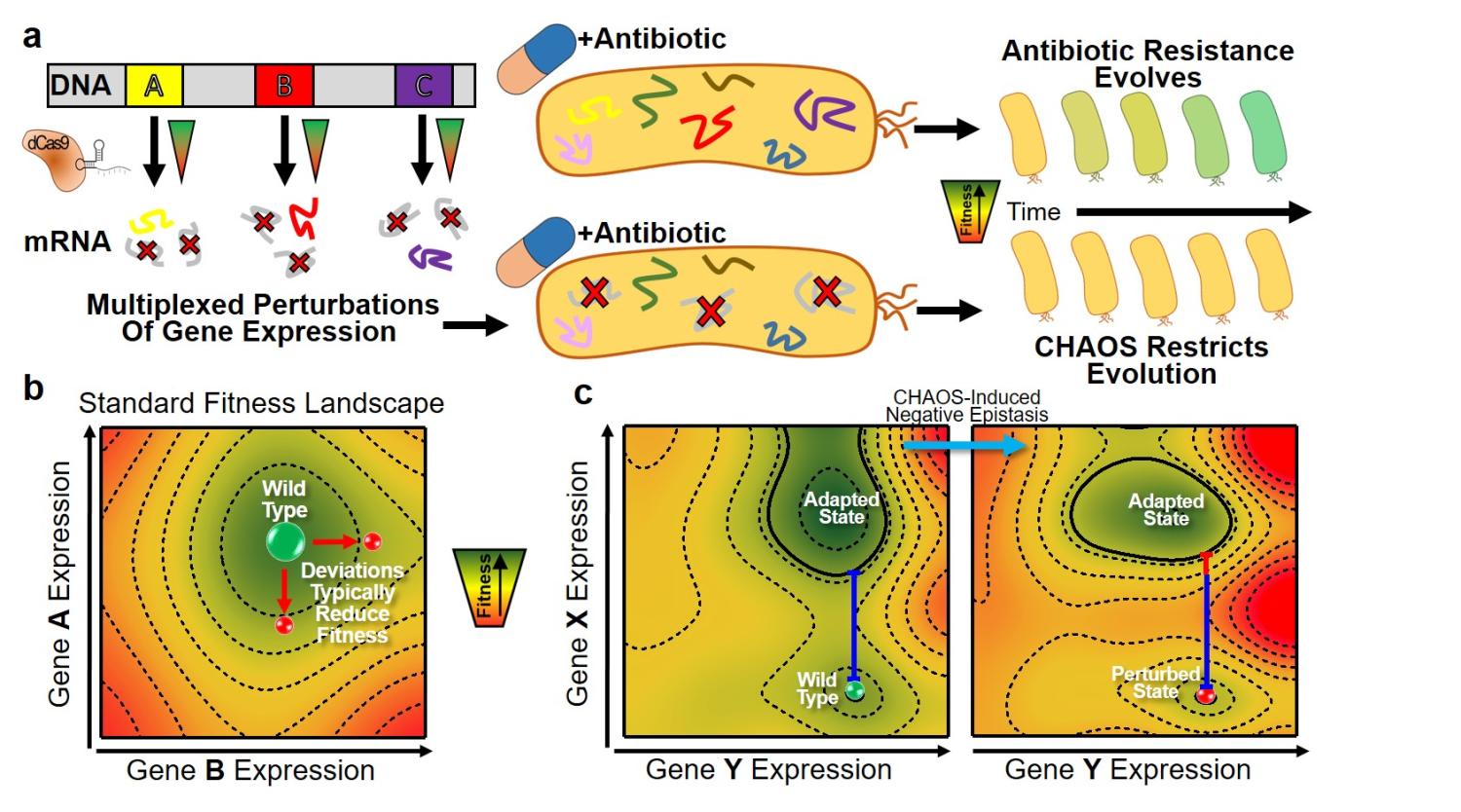
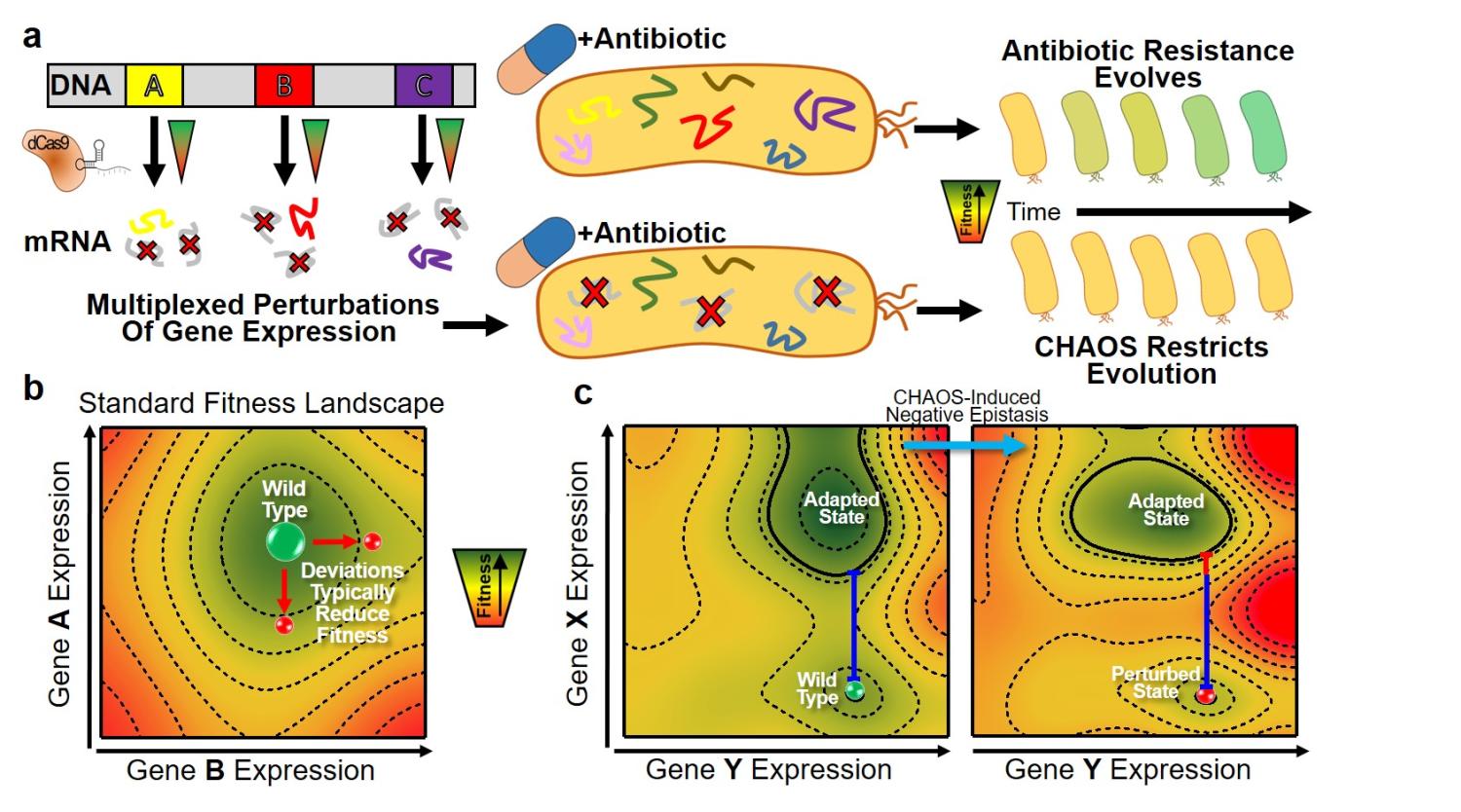
We report novel CHAOS Therapy to Slow down bacterial evolution of antibiotic resistance in paper "Multiplexed deactivated CRISPR-Cas9 gene expression perturbations deter bacterial adaptation by inducing negative epistasis" published in Nature Communications Biology.
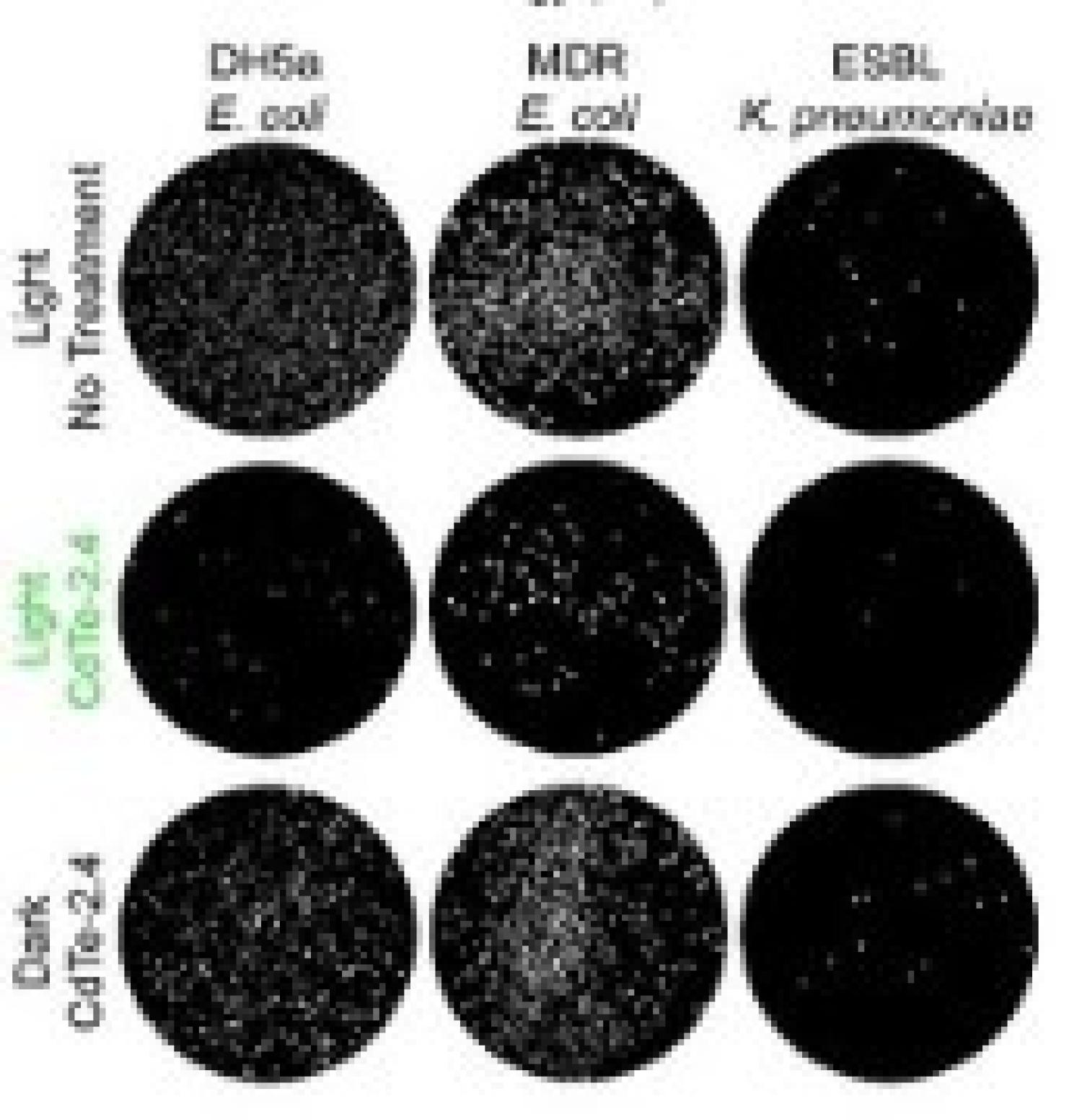
We report using Quantum dot antibiotics to potentiate antibiotics in highly drug-resistant clinical isolates in paper "Potentiating clinical drug resistant bacteria via stimuli-activated superoxide generation" published in Science Advances.
We report a novel Light-Activated Nanotherapy that kills Clinically isolated Multi Drug Resistant Bacteria in manuscript led by Colleen and Sam, "Photoexcited Quantum Dots for Killing Multi-Drug Resistant Bacteria" in Nature Materials!
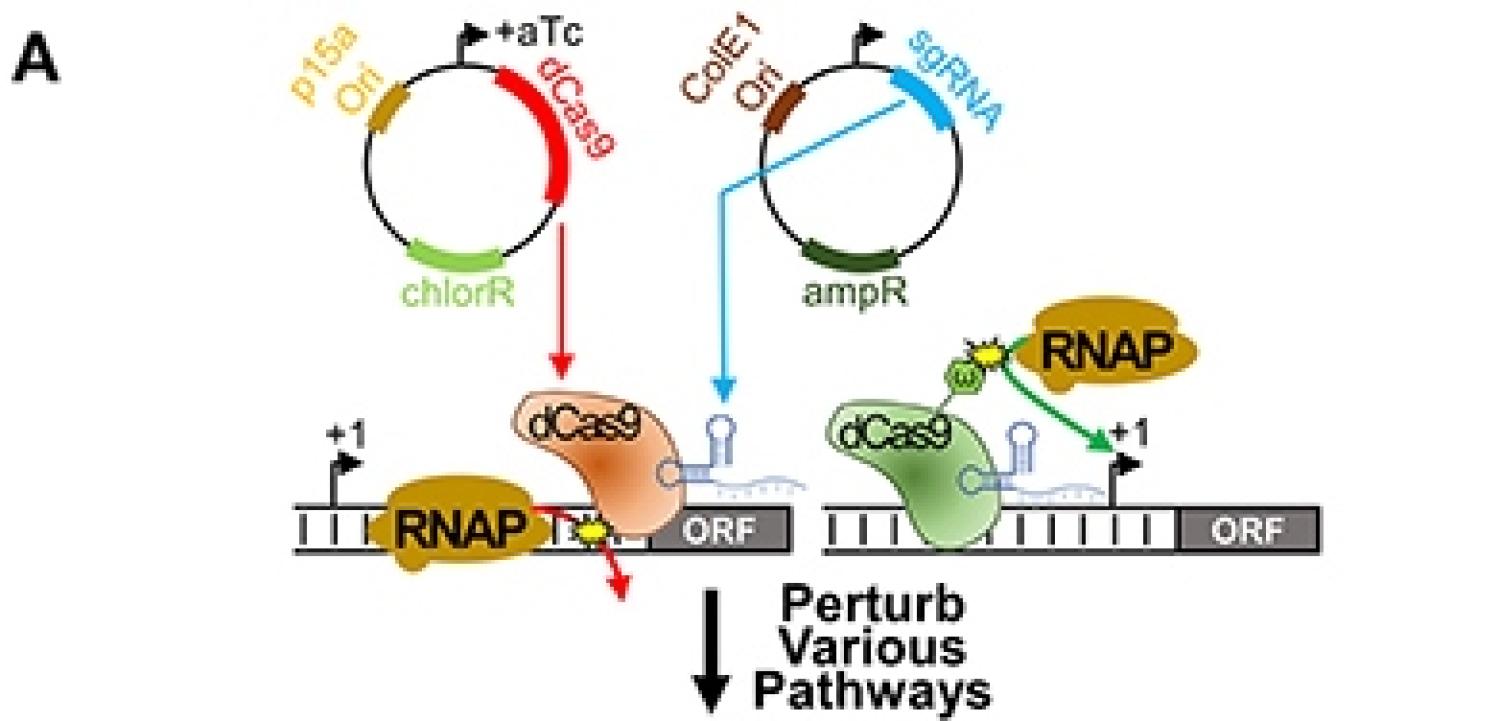
We report a CRISPR gene expression perturbation approach to increase bacterial tolerance to biofuels "CRISPR gene expression perturbations for enhancing bacterial tolerance to biofuels" in Frontiers in Bioengineering and Biotechnology!
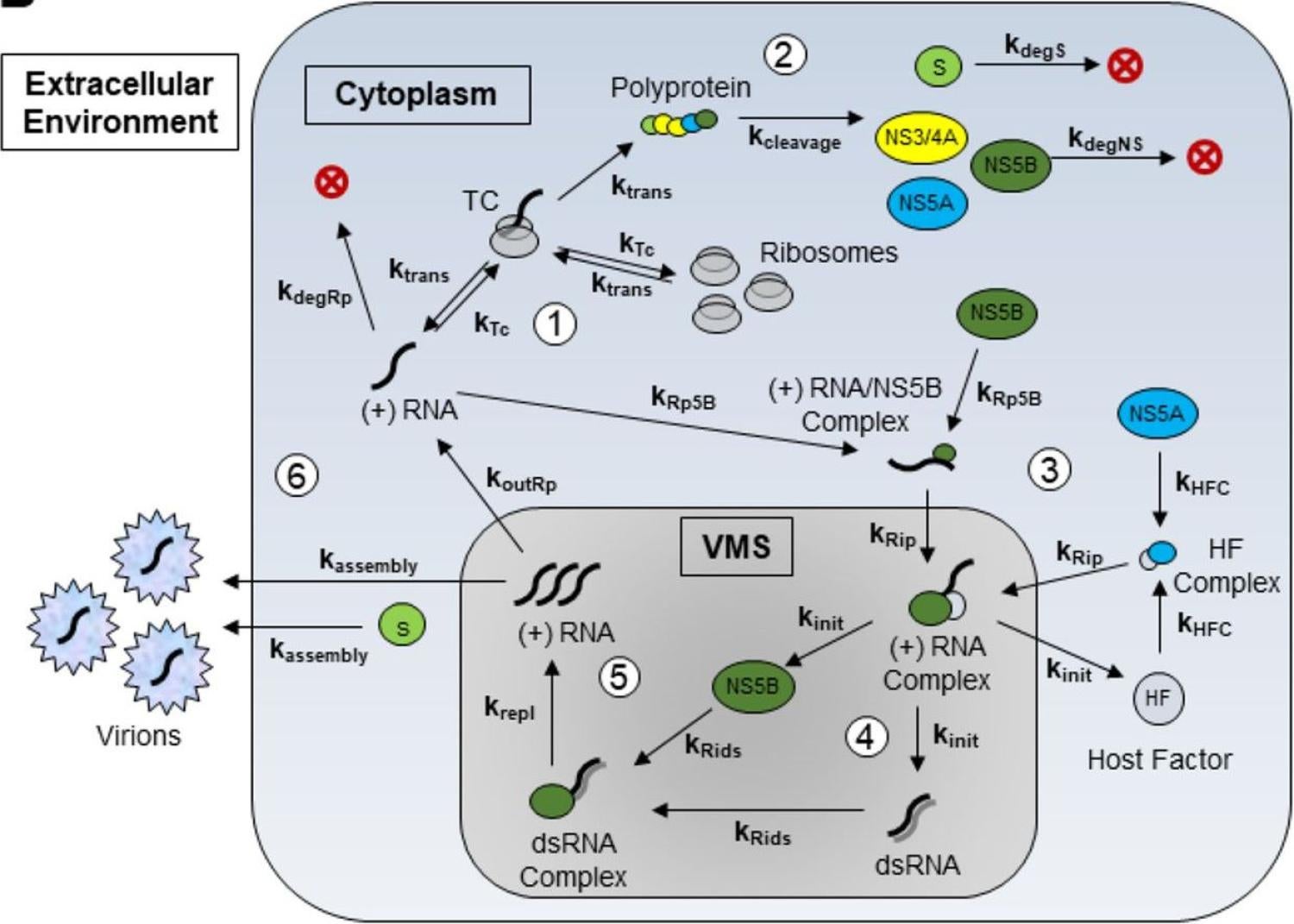
We report a new mathematical model fitted to experimental data for predicting intracellular HCV dynamics, "Intracellular hepatitis C modeling predicts infection dynamics and viral protein mechanisms" in Journal of Virology!
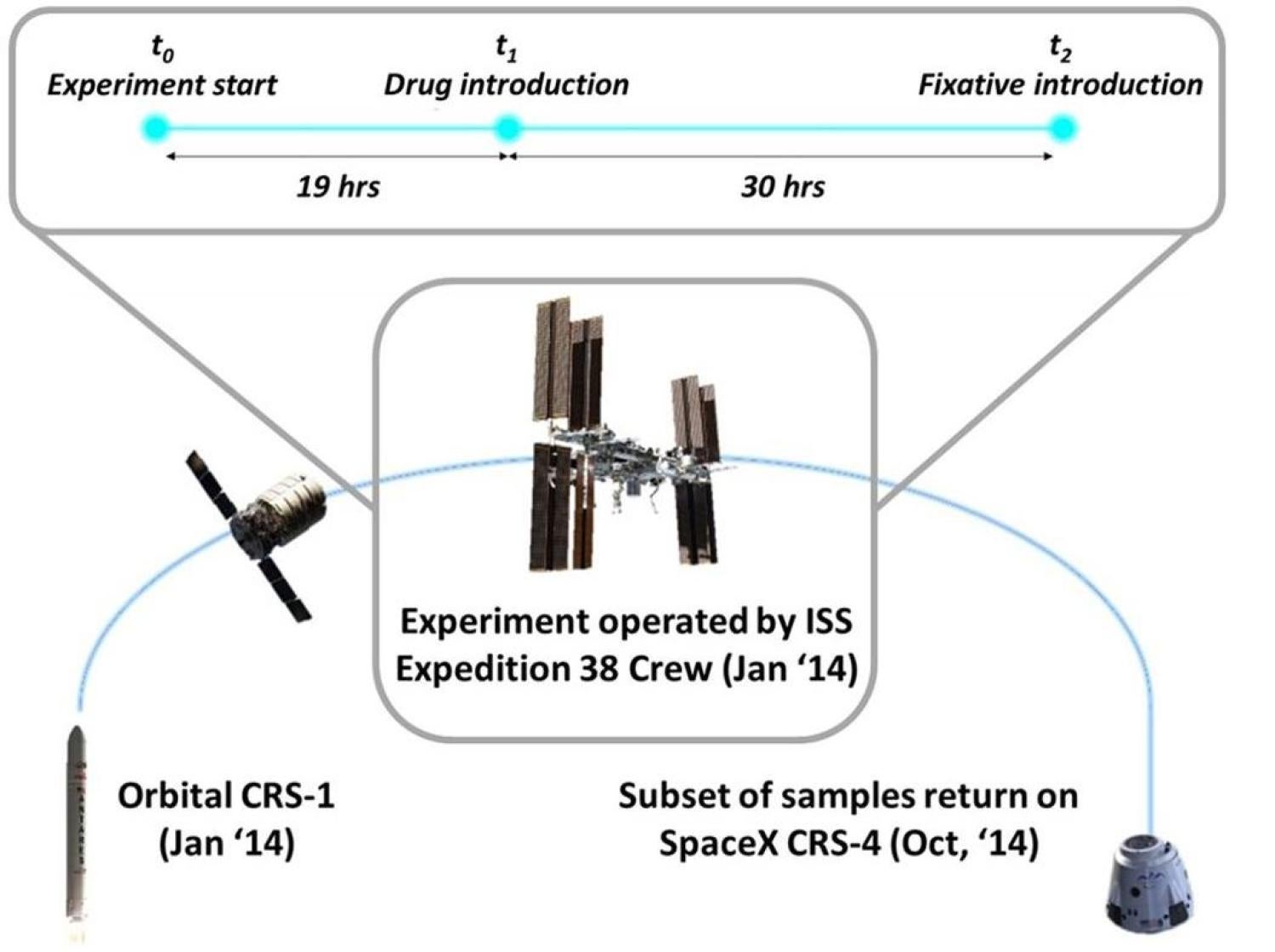
We report transcriptomic signature of E. coli adapted to antibiotics on the International Space Station, in "Spaceflight modifies Escherichia coli gene expression in response to antibiotic exposure and reveals role of oxidative stress." Frontiers of Microbiology!
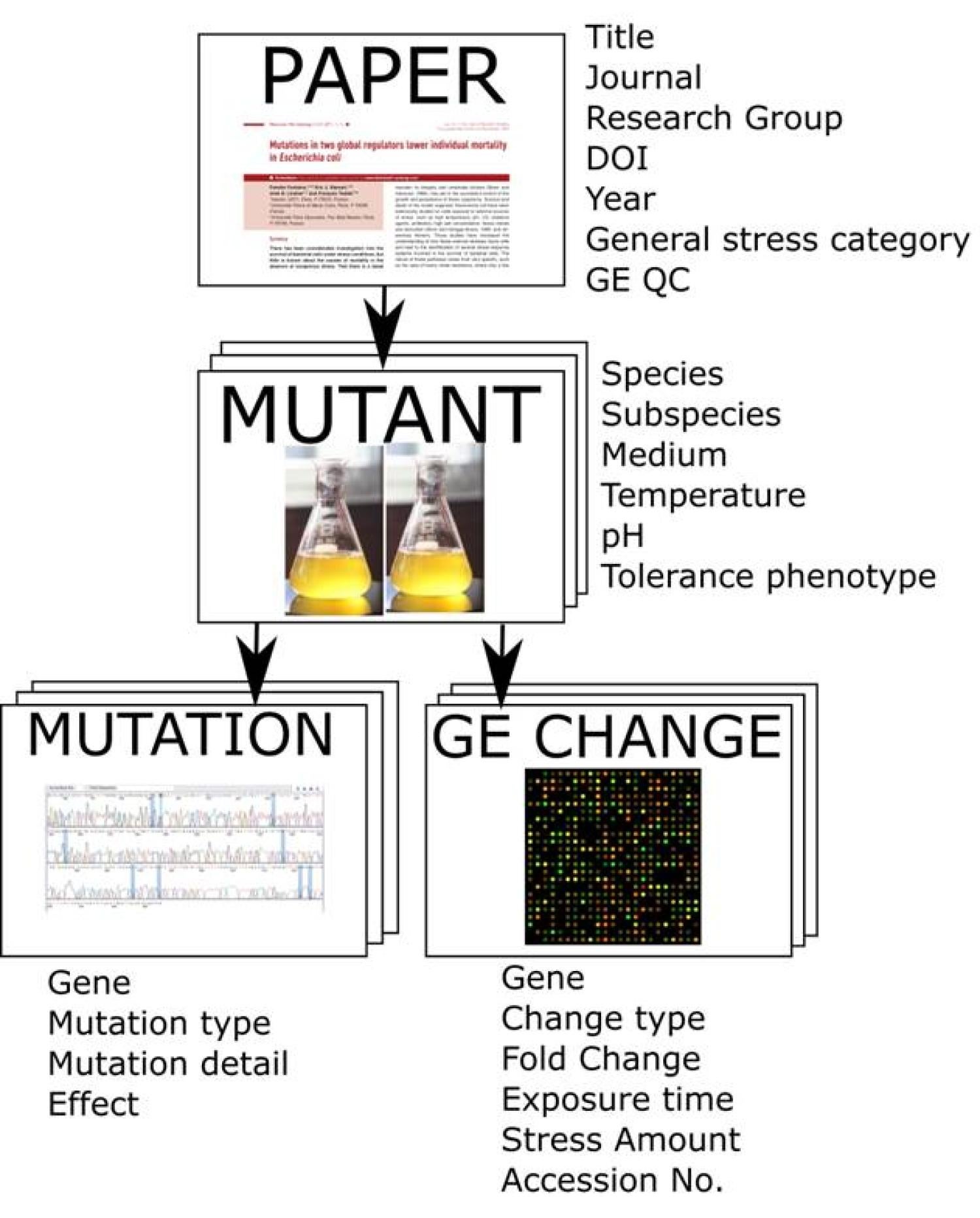
We report the first database that can mine gene expression data sets to identify key players of adaptive resistance in bacteria. "The Tolerome: A database of transcriptome-level contributions to diverse Escherichia coli resistance and tolerance phenotypes." published in ACS Synthetic Biology.
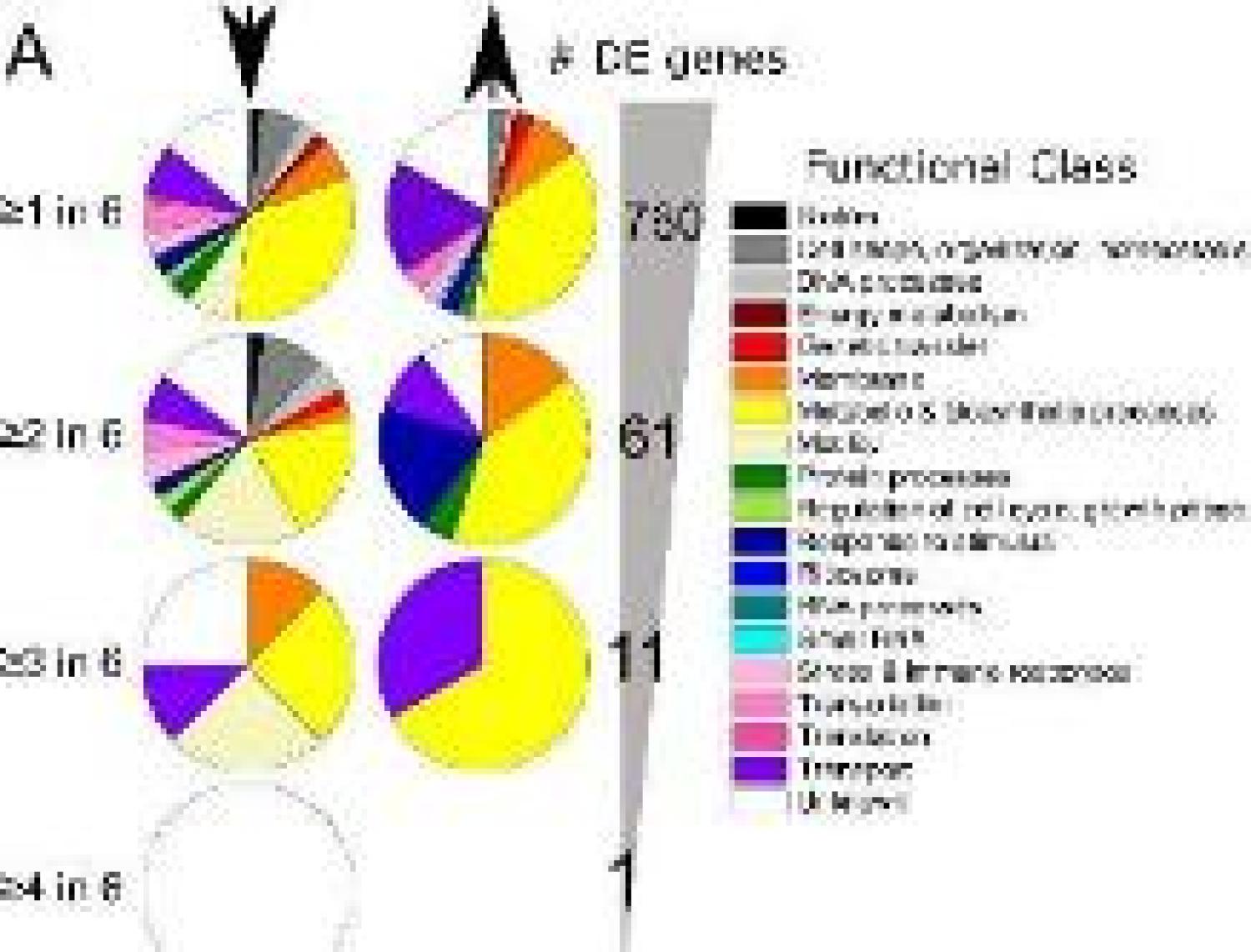
We are pleased to report a new systems bology approach to mine and identify key gene signatures in complex gene expression data sets in our publication "Transcriptome level signatures in gene expression and gene expression variability during bacterial adaptive evolution," in mSphere.
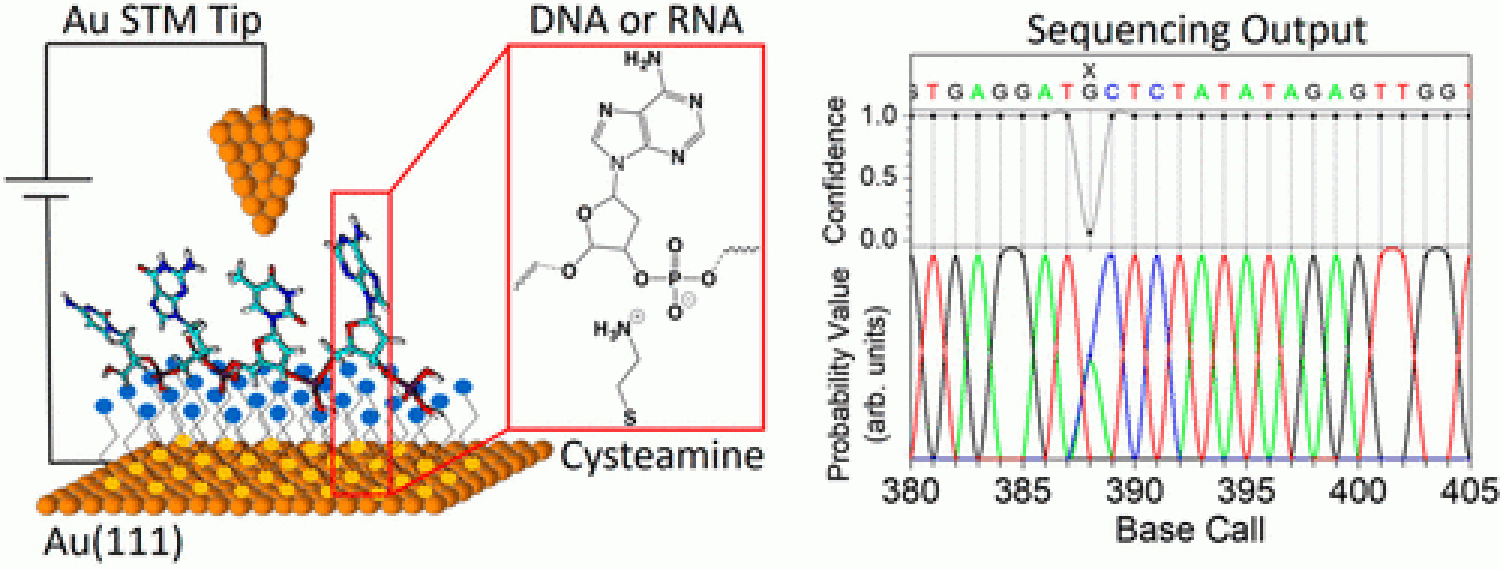
We are pleased to report four publications on novel single-molecule sequencing methods in 2017! These publications can be found in ACS Nano, JACS, and Small.
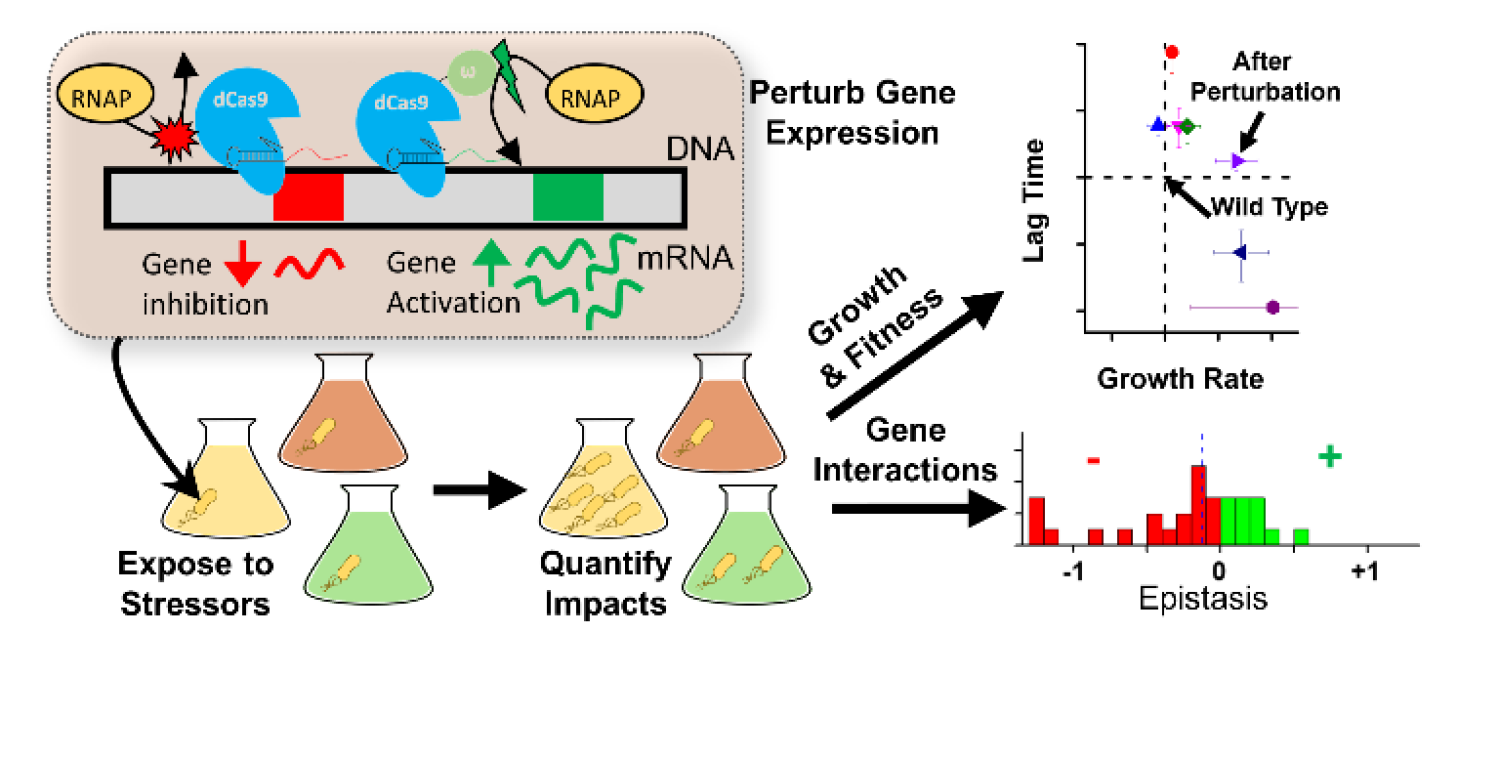
We present a novel application of CRISPR-Cas9 technology for investigating how gene expression governs the adaptive pathways available to bacteria during the evolution of resistance in Peter's paper titled, "CRISPR perturbation of gene expression alters bacterial fitness under stress and reveals underlying epistatic constraints" in ACS Synthetic biology!
We report a novel tool for synthetic biology and metabolic engineering by investigating transcriptional interference mechanisms of antisense roadblock and RNA polymerase traffic in a set of convergent promoters, in paper led by Toni, "Transcriptional Interference in convergent promoters as a means for Tunable Gene Expression" in ACS Synthetic biology!
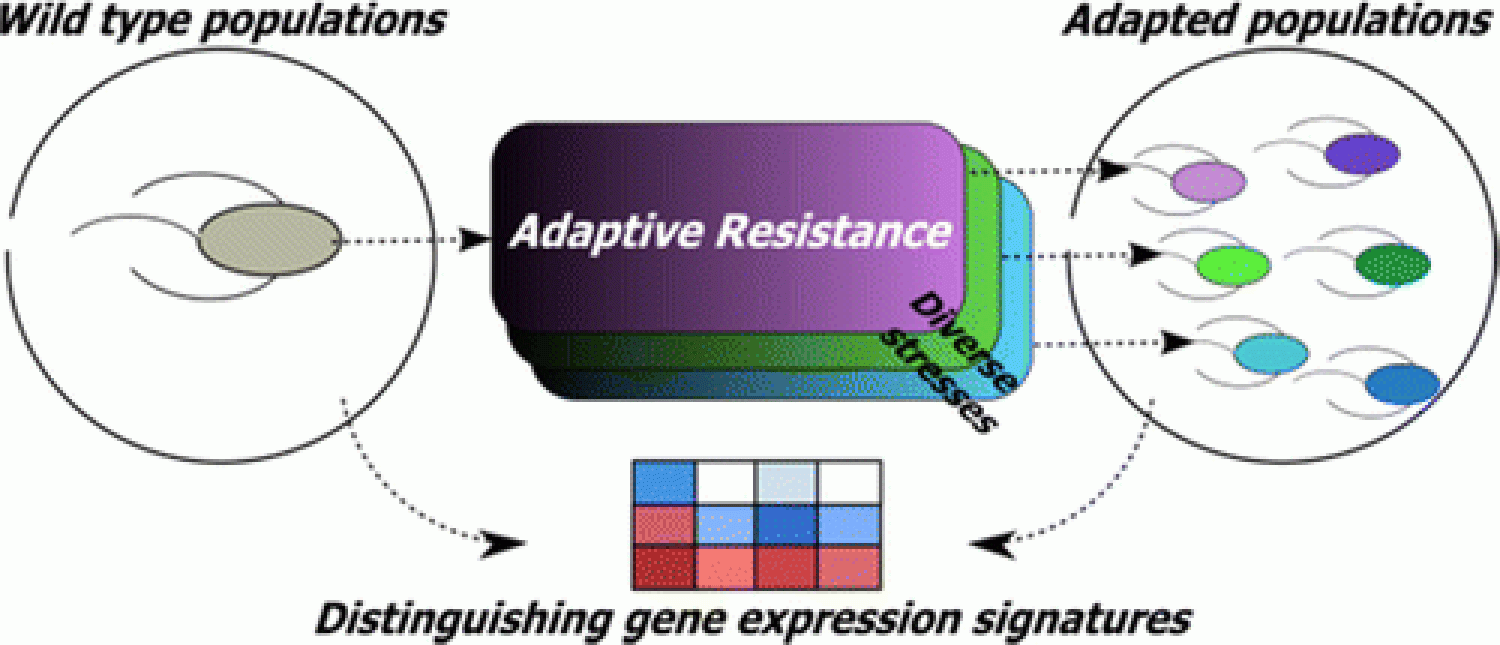
We report a novel gene expression variability based approach to identify important genes involved in bacterial adaptive resistance, in paper led by Keesha, "Gene expression variability underlies adaptive resistance in phenotypically heterogenous bacterial populations" in ACS Infectious Diseases!
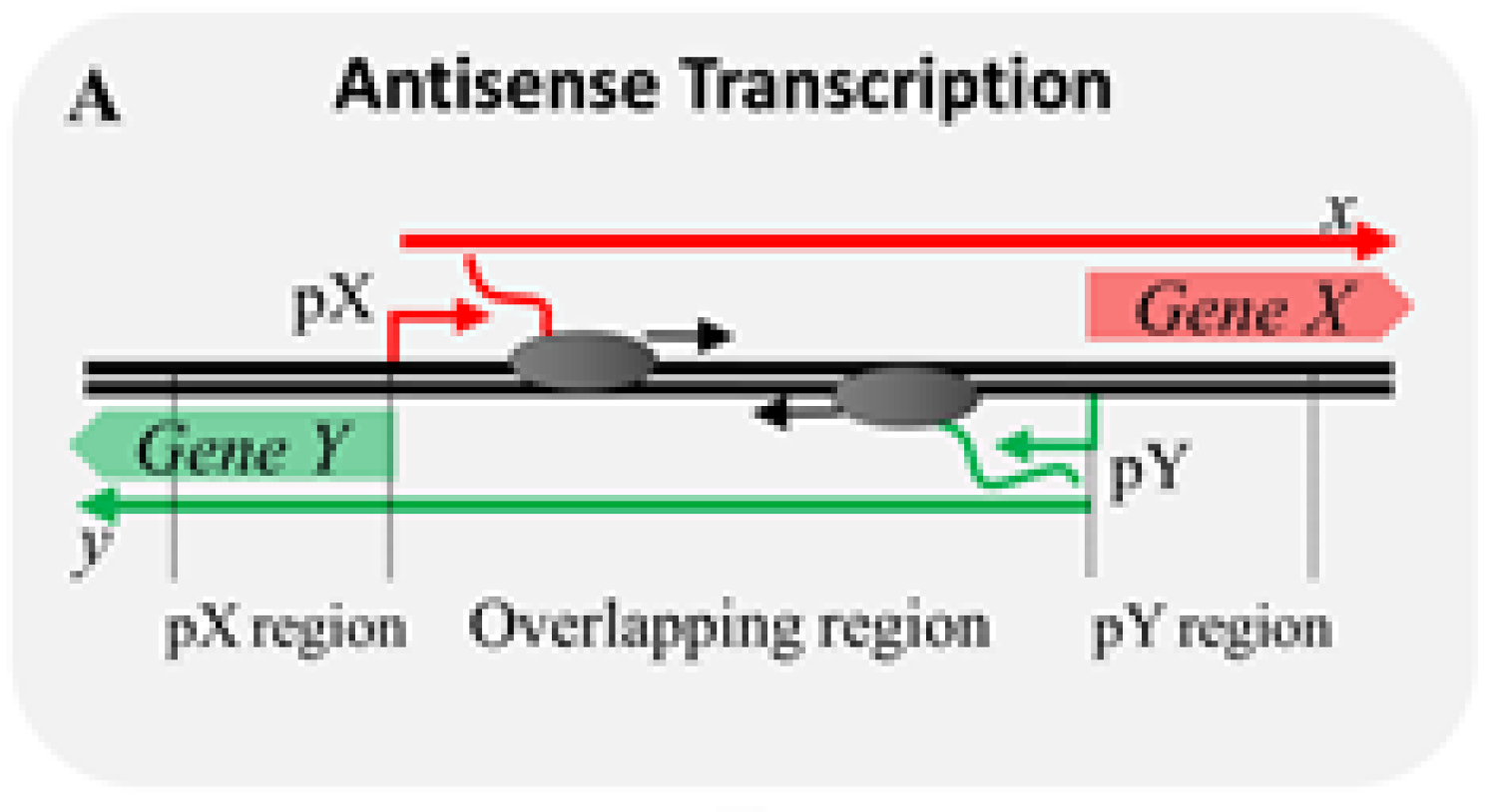
We report a new mathematical model that describes tunabilty of genetic switch behavior during antisense transcription, in a paper led by Toni, "cis-Antisense Transcription Gives Rise to a Tunable Genetic Switch Behavior: A Mathematical Modeling Approach" in PLoS ONE!
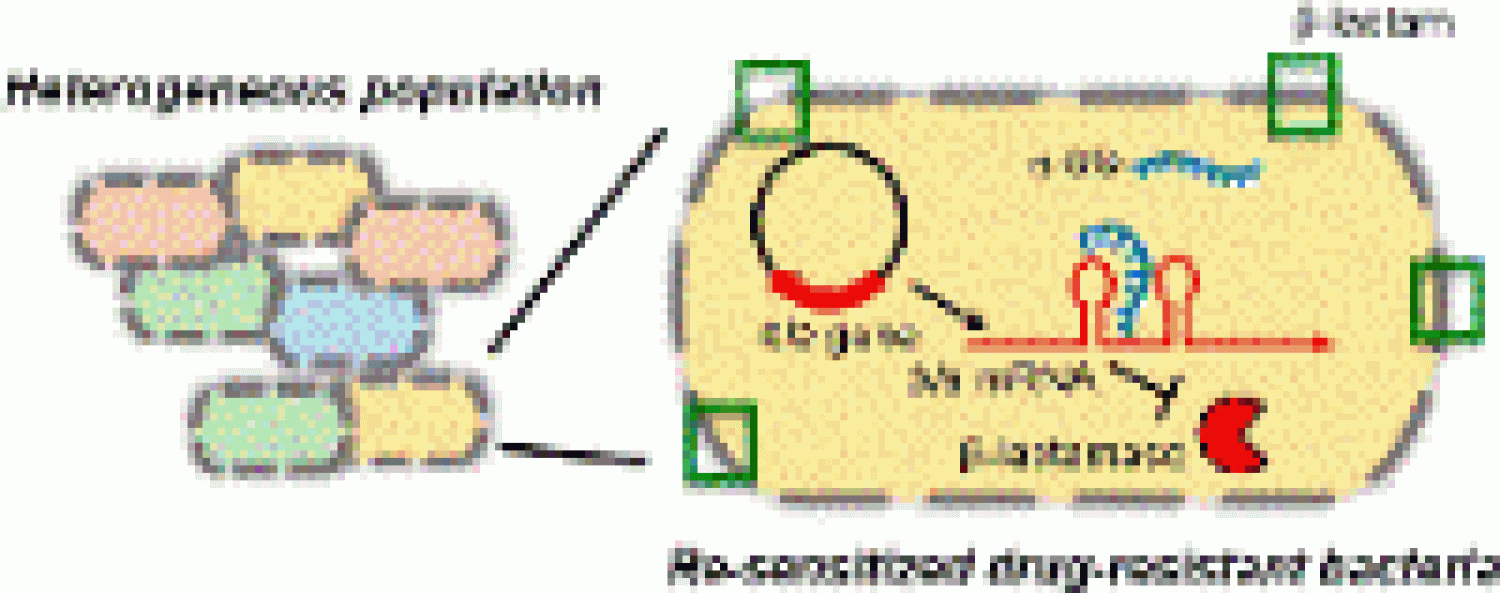
We report a bet-hedging based mechanism of adaptive resistance to PNA based antimicrobials in a paper led by Colleen, "Sequence-specific peptide nucleic acid based antisense inhibitors of TEM-1 beta-lactamase and mechanism of adaptive resistance" in ACS Infectious Diseases!
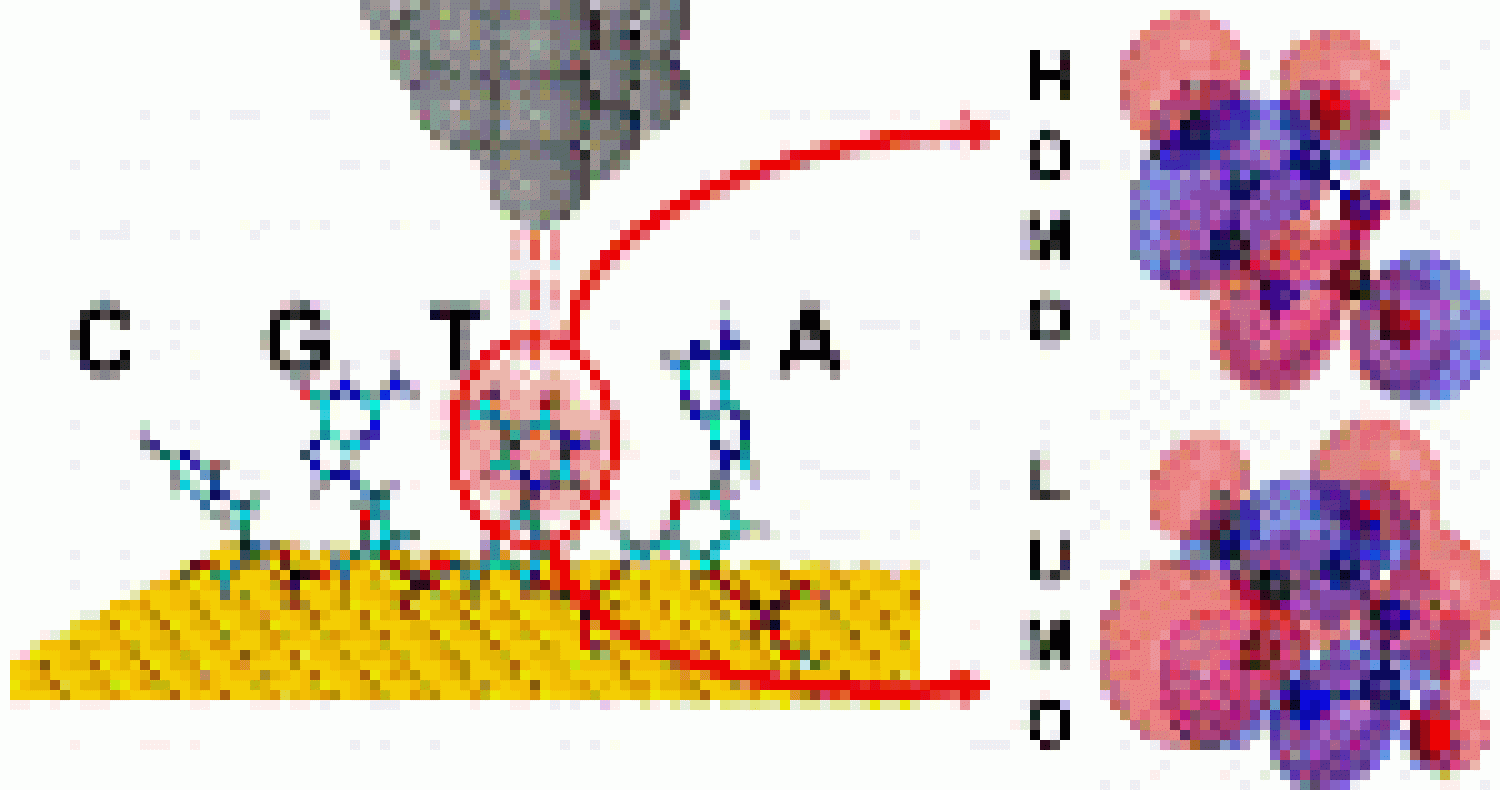
We reveal novel nanoelectronic fingerprints for DNA in paper led by Josep, "Measurements of Single nucleotide Electronic states as Nanoelectronic fingerprints for Identification of DNA Nucleobases, their Protonated and Unprotonated states, Isomers and Tautomers," in Journal of Physical Chemistry Letters B!
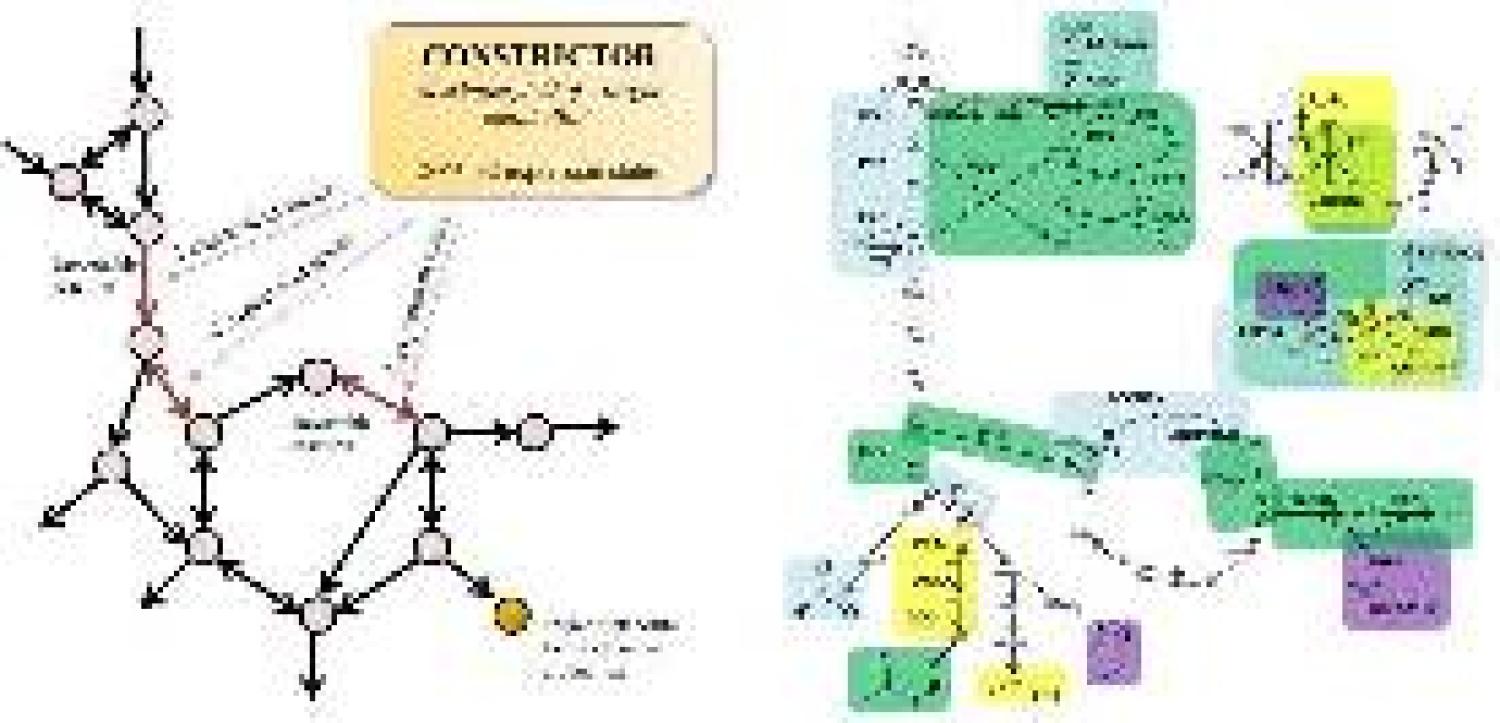
New Metabolic Flux Analysis tool CONSTRICTOR is available on Opensource
Our lab designs and reports a new FBA tool called CONSTRICTOR, in a paper led by Keesha, "CONSTRICTOR: Constraint modification provides insight into design of biochemical networks" in PLoS ONE!
Sam's paper "Multiple Energy Exciton shelves in Quantum Dot-DNA Nano-bioelectronics" is published in The Journal of Physical Chemistry Letters!
Our mathematical modeling paper on "A pharmacokinetic/viral kinetic model to evaluate treatment of chronic hepatitis C virus with danoprevir" accepted in Antiviral Therapy!
Anushree's paper on "Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer" is published in PNAS!

Welcome
Our research group uses a combination of interdisciplinary approaches including synthetic biology, systems biology, microbiology, synthetic chemistry, and nanoengineering to address the key global challenge involving antimicrobial resistance. We aim at developing strategies for rationally engineering next-generation smart antimicrobials that can eliminate multi-drug resistant superbugs, with the ultimate goal of creating therapies that can evade evolution of drug-resistance in microbes. To this end, we are also interested in investigating the non-genetic mechanisms responsible for antimicrobial tolerance/resistance to devise strategies that can one day prevent evolution of drug resistance. Using synthetic biology tools we are also interested in designing, constructing and engineering modular synthetic genetic devices based on less-explored naturally occurring mechanisms, for both understanding fundamentally how nature works as well as built devices for potential biotechnological applications.
Featured by CU Engineering
[video:https://youtu.be/VcXuTGNphPY]
Scientists are using chaos to try and beat E. coli
--by Chase Purdy, Quartz
For years, scientists at the University of Colorado have sought to defuse the destructive powers of a pernicious bacteria that often sneaks into the human body through tainted food. Turns out, one of the most effective ways to beat E. coli is to sow chaos. (Read more)
Lab invents CHAOS Therapy to Slow down the bacterial evolution of antibiotic resistance
How to stop an antibiotic-resistant superbug
-- By Trent Knoss, University of Colorado press release
A genetic disruption strategy developed by University of Colorado Boulder researchers effectively stymies the evolution of antibiotic-resistant bacteria such as E. coli, giving scientists a crucial leg up in the ongoing battle against deadly superbugs. (Read more)
Research led by CU Boulder could block E.coli, other superbugs
-by CHARLIE BRENNAN, Denver Post and Boulder Daily Camera
Kryptonite for superbugs could be on the horizon, as a result of research led by University of Colorado researchers that hinders the progress of antibiotic-resistant superbugs such as E.coli... (Read more)
Anushree speaks at the Institute of Systems Biology's Future of Health Symposium
[video:https://youtu.be/wB5Q9gqMMcw]
Session 3: Prevention and Therapy Anushree Chatterjee, PhD | University of Colorado at Boulder "From Synthetic Biology to Nanobiotechnology: Rational Antimicrobial Engineering Approaches for Combating Multidrug-Resistant Pathogens"
BBC features our Quantum dot antibiotic research!
CGTN America features our lab's research
[video:https://youtu.be/VSE2RRv4JEc]
Battling Bacteria: Back to Square 1?
--Lab Research Featured by Hamodia
A WATERSHED IN RESEARCH STUDIES
Trailblazing research studies of diverse approaches, recently published in peer-reviewed science journals, are poised to combat antibiotic resistance. Hamodia interviewed these leading researchers, each at the helm of his study. Assistant Professors Prashant Nagpal and Anushree Chatterjee of the Department of Chemical and Biological Engineering in the University of Colorado Boulder, whose studies were published in the science journals Nature Materials and Science Advances. “Our approach is a two-pronged strategy to fight antibiotic-drug resistance..... (Read more)
MSN, Newsweek, Science News, CIDRAP features our work on potentiating drug-resistant clinical bacteria to antibiotics
[video:https://youtu.be/qBCrZCmgZos]
Boulder, Colorado —Light-activated nanoparticles, also known as quantum dots, can provide a crucial boost in effectiveness for antibiotic treatments used to combat drug-resistant superbugs such as E. coli and Salmonella, new CU Boulder research shows......
UC Health features our research on combating antibiotic resistance
Todd Neff for UC Health- DENVER, CO. Science can border on science fiction. Designing swarms of infinitesimal semiconductors that, shined with green light, turn standard oxygen into a superoxide that makes life hell for drug-resistant bacteria – that qualifies.....
Voice of America features our Quantum dot based Antibiotic research
[video:https://youtu.be/6xGn-z6B_uM]
Boulder, Colorado — Scientists have been creating antibiotics to kill deadly bacteria for nearly 80 years. Unfortunately, the bacteria that cause disease can adapt so quickly, some have evolved into superbugs that are resistant to all antibiotics, and making new drugs is a long, expensive, involved process....
ABC Channel News Features our Light-activated nanotherapy against antibiotic-resistant “superbugs”!
[video: https://www.thedenverchannel.com/news/local-news/cu-researchers-use-light-activated-nanoparticles-used-to-kill-antibiotic-resistant-superbugs]
In the ever-escalating evolutionary battle with drug-resistant bacteria, humans may soon have a leg up thanks to adaptive, light-activated nanotherapy developed by researchers at the University of Colorado Boulder.
Antibiotic-resistant bacteria such as Salmonella, E. Coli and Staphylococcus infect some 2 million people and kill at least 23,000 people in the United States each year. Efforts to thwart these so-called “superbugs” have consistently fallen short due to the bacteria’s ability to rapidly adapt and develop immunity to common antibiotics such as penicillin.
New research from CU-Boulder, however, suggests that the solution to this big global problem might be to think small—very small.
In findings published today in the journal Nature Materials, researchers at the Department of Chemical and Biological Engineering and the BioFrontiers Institute describe new light-activated therapeutic nanoparticles known as “quantum dots.” The dots, which are about 20,000 times smaller than a human hair and resemble the tiny semiconductors used in consumer electronics, successfully killed 92 percent of drug-resistant bacterial cells in a lab-grown culture.....
News
Shane and Yuchen's paper "Light-driven Metabolic Pathways in Non-photosynthetic Biohybrid Bacteria" published in ChemBioChem. Congratulations!
Christian's paper "High Throughput Viability Assay for Microbiology" accepted in Nature Microbiology. Congratulations!
Check out how to use the Geometric Viabilty Assay for speeding up your microbiology research using the link provided.
[video:https://www.youtube.com/watch?v=9tAIbpZ_uOw]
Yuchen's paper Light-driven Transformation of Carbon Monoxide into Hydrocrbons using CdS@ZnS:VFe Protein Biohybrids published in ChemSuSChem. Congratulations!
Christian awarded NIH K99 Pathway to Independence award! Congratulations!
Fateh and Yangming complete a successul summer internship in the Chatterjee lab!

Welcome Fateh to the lab for antibitic research! Thanks to support from Mitacs Globalink Research Award Abroad.
Safety and biodistribution study for COVID targeting direct-acting antiviral is now published in ACS Biomaterial Science and Engineering!
Congratulations Cory for winning the Advanced Light Source (ALS) Doctoral Fellowship at Berkeley Lab!
Congratulations Christian on being awarded the Fall 2022 PAC Research/Outreach Grant!
"Photoactivated antibiotics to treat intracellular infection of bacteria" on the use of QD antibiotics to reduce intracellular infections in a bone-infection model is published in Nanoscale Advances!
Christian Meyer joins lab as a post-doc. He brings his expertise in biophysics, drug-drug interactions, as well as novel high-througput methods to measure bacterial viability! Welcome!
Nolan O'Connor defends Ph.D. and becomes Dr. O'Connor! Congratulations!!
Colleen McCollum defends Ph.D. and becomes Dr. McCollum! Congratulations!!

Kristen Eller defends Ph.D. and becomes Dr. Kristen Eller! Congratulations!!
Peter and Kristen's paper on "Potentiating antibiotic treatment using fitness-neutral gene expression perturbations" published in Nature Communications Biology.
"Role of miR-2392 in driving SARS-CoV-2 infection," an international collaborative effort to develop treatment against SARS-CoV2 published in Cell Reports.
"Therapeutics in less than a week." Our Nature Comm Biology paper got selected for an article in The Conversation US. Congrats Kristen! Check it out !
FAST paper recieves press: Eurek Alert | Science Daily | PhyS Org | Bio IT World | Bioengineer org | ScienMag | Mirage News | Technology Networks | CU Boulder | Boulder Daily Camera | The Tribune | MSN News | The Conversation | Japan Today | Yahoo | Buffalo News | Foreign Affairs
Tom Aunins becomes Dr. Thomas Aunins and graduates with a Ph.D. Congratulations!! Thanks for all the great work and best of luck for the next chapter of your career!
"Feeling the need for speed (for new therapeutic modalities). Outsmarting evolving microbes." Check out our new paper on translation of Quantum dot antibiotics: Colleen R. McCollum, John R. Bertram, Prashant Nagpal, and Anushree Chatterjee* (2021). Photoactivated indium phosphide quantum dots treat multi-drug resistant bacterial abscesses in vivo. ACS Applied Materials & Interfaces (In press).
- Dana's paper on "Light-Activated Quantum Dot Potentiation of Antibiotics to Treat Drug-Resistant Bacterial Biofilms" published in Nanoscale Advances
- Shane recieves NSF GRFP Honorable mention! Congrats!
- "Feel the need for speed?" FAST Antimicrobials in less than one week. Kristen's paper on "Facile accelerated specific therapeutic (FAST) platform develops antisense therapies to counter multidrug-resistant bacteria" published in Communications Biology
- Nolan's paper "Engineering Transcriptional Interference through RNA Polymerase Processivity Control" published in ACS Synthetic Biology
- Colleen's paper on Quantum dot antibiotic animal safety and efficacy study "Photoexcited quantum dots as efficacious and nontoxic antibiotics in an animal model" published in ACS Biomaterials Science & Engineering
- Tom and Keesha's paper on "Transcriptome-based design of antisense inhibitors potentiates carbapenem efficacy in CRE E. coli" published in PNAS
- Lab recieves funding from NASA-TRISH to create technology to mitigate the effects of space radiation as part of an interdisciplinary team led by CSU
- Lab featured on American Physical Society News
- Lab collaborates on LDRD project with Sandia National Laboratory
- Lab recieves funding from The Translational Research Institute for Space Health (TRISH) to support COVID-19 research
- Lab supports COVID-19 International Research Team (COV-IRT) in search of therapeutic solutions against SARS-CoV2
- ARMOR releases podcast series on Spotify: ARMOR on the air
- Lab featured on On CUE Podcast: The Anti-Microbial Resistance Mediation Outreach Program (ARMOR)
- Lab outreach effort ARMOR featured on cover of CUE Engineering Magazine
- Lee defends Ph. D. thesis virtually and becomes Dr. Korshoj! Many congratulations on a job welldone!
- Lab interviewed about COVID-19
- 'Outsmarting pathogens' using the FAST platform - American Physics Society (APS) press
- We are proud to present the first Antimicrobial Regeneration Consortium (ARC) seminar by Dr. John Aunins on Feb 10th, 2020!! The seminar will be livestreamed globally. Watch Dr. Aunins' Seminar Here!

- Lab recieves NIH Division of Microbiology and Infectious Diseases (DMID) contract for studying efficacy of QD antibiotics in animal models.
- Super FAST response to disease outbreaks: Lab featured as Leading Edge in CU Engineering Magazine Fall 2019 edition.
- Toni and Nolan's paper on Construction of two-input logic gates using Transcriptional Interference published in ACS Synthetic Biology.
- Lab receives a $2.25M Department of Energy grant led by the Nagpal lab to develop a quantum dot toolkit for multimodal bioimaging
- Lab receives funding from Vastbiome Inc. to study gut bacteria.
- Anushree recieves Provost Faculty Achievement Award
- Lab issued two patents: Quantum Dot Antibiotics (US Patent: 10383939) and Nanoelectronic sequencing (US Patent: 10364461)
- Tom's paper on "Isolating the Escherichia coli transcriptomic response to superoxide generation from cadmium chalcogenide quantum dots," published in ACS Biomaterials Science and Engineering.
- Dr. Sadhana Sharma joins lab as a research assistant professor/senior postodoctoral fellow. Welcome!
- Two of Max's papers published. Near-infrared light triggered antimicrobial indium phosphide quantum dots paper published in Angewandte Chemie and Tuning ternary Zn1-xCdxTe quantum dot composition: Engineering electronic states for light-activated superoxide generation as a therapeutic against multidrug-resistant bacteria in ACS Biomaterials and Engineering!
- Anushree receives Outstanding Junior Faculty Award from ChBE!
- ARMOR program in full flow! Read more: https://www.co-arc.org/news-updates/cu-students-teach-underrepresented-high-school-students-about-antimicrobial-resistance
- Congrats to high school student Anudeep Golla for winning 2nd out of around 60 projects in Computational Biology catgory at ISEF! He won $$ and was chosen to have an asteroid named after him!
- Antimicrobial Resistance Mediation OutReach (ARMOR) Program CU Boulder Chapter official website launched! See link. Check out our outreach events!
- Lab undergraduate researcher Vismaya Bachu wins Silver Medal Award from the College of Engineering! Congratulations!! :) Vismaya will be heading to Johns Hopkins medical school this fall.
- REU student Suxuen Yew from Kansas University joins lab for summer research. Welcome!
- New Postdoc Job posting: See here
- Lab receives funding from NASA-TRISH and Lab Venture Challenge!
- Anushree wins Lab Venture challenge award for developing novel nucleic acid therapies against Multi-drug resistant bacteria.
- Lab develops a de-novo aggregating antimicrobial peptide and bacterial conjugation based delivery system.
- Coordinated CHAOS: CU Science Buffs blog about CHAOS Therapy
- Denver Post highlights CHAOS therapy research
- CHAOS paper receives significant media press: University of Colorado press release, Phys.Org, Scicasts, Science Daily, Innovation News, Quartz, and more.
- Toni successfully defends his Ph. D. and becomes Dr. Antoni E. Bordoy! Many congratulations!!
- Peter's paper "CRISPR gene expression perturbations for enhancing bacterial tolerance to biofuels" published in Frontiers in Biotechnology and Bioengineering. Congrats!
- Peter successfully defends his Ph. D. and becomes Dr. Peter Otoupal! Peter will move on to his new post-doc position at Sandia National Lab starting in August. Many congratulations Peter!
- Peter's paper "Multiplexed deactivated CRISPR-Cas9 gene expression perturbations deter bacterial adaptation by inducing negative epistasis" is published in Nature Communications Biology! Congrats!
- Anushree nominated as "Rising Stars" by the journal Frontiers of Chemistry!
- Peter wins Excellence in Graduate student research award from ChBE department. Congrats!
- Will Cordell wins Marilyn and Howard L. Anseth Outstanding Undergraduate Research Award and Outstanding Undergraduate Research Awards ‐Gold medal. Congrats!
- Kristen and Tom recieve NSF GRFP Honorable mentions. Congrats!
- Anushree receives ACS Infectious Diseases Young Investigator Award!
- Anushree becomes Associate Editor of Frontiers in Bioengineering and Biotechnology.
- BBC features our Quantum dot antibiotic research! Link to article
- Tom's paper "Intracellular hepatitis C modeling predicts infection dynamics and viral protein mechanisms" published in Journal of Virology. Congrats Tom!
- Tom's paper "Spaceflight modifiesEscherichia coli gene expression in response to antibiotic exposure and reveals role of oxidative stress" is published in Frontiers in Microbiology. Congrats Tom!
- University of Colorado Health features our research to combat antibiotic resistance! Link to article.
- Peter wins third place in Stars symposium. Congrats!
- Lee's paper on "Conformational Smear Characterization and Binning of Single-Molecule Conductance Measurements for Enhanced Molecular Recognition" published in JACS. Congratulations!
- Keesha's paper on "The Tolerome: A database of transcriptome-level contributions to diverse Escherichia coli resistance and tolerance phenotypes" published in ACS Syn Bio. Congratulations!
- Our potentiating antibiotics paper in Science Advances receives press coverage including
- Colleen's paper on "Potentiating clinical drug resistant bacteria via stimuli-activated superoxide generation" published in Science Advances. Congratulations!
- Sepideh's paper "Quantum Point Contact Single-Nucleotide Conductance for DNA and RNA Sequence Identification" published in ACS Nano. Congratulations!
- Colleen moves on to her new post-doc position at Sandia National Lab at Livermore. Congratulations and all the best!
- Anushree receives grant funding from National Science Foundation!
- Colleen successfully defends her Ph.D., and becomes Dr. Courtney!! Congratulations!!
- Colleen recieves Max Peters Outstanding Graduate Student Award. Congratulations!
- Keesha successfully defends her Ph.D. and becomes Dr. Erickson! Keesha moves on to pursue postdoctoral research at Los Alamos National Laboratory. Congratulations and all the best!!
- Anushree recieves DARPA Young Faculty Award! Link
- Lee wins NSF Graduate Fellowship. Congratulations!
- Keesha's paper on "Transcriptome level signatures in gene expression and gene expression variability during bacterial adaptive evolution”published in mSphere. Congrats!
- Voice of America features our Quantum Dot Antibiotic work in article, "Tiny Robots May Someday Fight Superbugs Inside Our Bodies" Link
- Colleen won 2nd place at the 9th Annual Bionanotechnology Graduate Student Award Session at AIChE. Her award talk was titled “Photoexcited Quantum Dots Potentiate Antibiotic Activity in Multidrug-Resistant Bacteria”. Congrats Colleen! Link
- Lee's paper titled "Single nucleotide DNA sequence identification using biophysical signatures from nanoelectronic quantum tunneling" published in Small. Congrats Lee!
- Colleen wins Best Poster Award from the Biochemical Engineering Journaland the AIChE Food, Pharmaceutical and Bioengineering Division for her work titled, "Sequence Specific Antisense Inhibitors of Non-Traditional Antibiotic Pathways." Congrats Colleen! Link
- Anushree recognized as one of the most highly prolific authors for ACS Infectious Diseases!
- Sam Goodman defends his Ph.D. and moves on to a postdoctoral fellow position at the National Academy of Sciences. Congrats Sam!
- Peter's paper on "CRISPR perturbation of gene expression alters bacterial fitness under stress and reveals underlying epistatic constraints" is published in ACS Synthetic Biology. Congrats Peter!
- Lab's Quantum dot antibiotic research featured by reporter Shelley Schlender on 88.5 FM KGNU Science Show "How on Earth." See KGNU Link
- Hear The Podcast
- Toni's paper on "Transcriptional Interference in convergent promoters as a means for Tunable Gene Expression" is published in ACS Synthetic Biology. Congrats Toni!
- Quantum dot antibiotic work highlighted in MRS Bulletin!
- See MRS Bulletin Link
- Quantum dot antibiotic work featured in France's Tech 24 TV show!
- See Tech24 Link
- Vismaya and Pallavi recieve UROP awards to pursue research. Congrats!
- Keesha recieves NSF-EAPSI Fellowship to conduct collaborative research. Congrats Keesha!
- Lee recieves GAANN fellowship. Congrats!
- Our Light Activated Nanotherapy featured in ABC's Denver Channel 7 News! Link to video
- Sepideh and Sajida join lab as postdoctoral fellows. Welcome!
- New graduate students Lee, Partha join Chatterjee Lab. Welcome!
- Keesha wins Dean's Graduate Student Research Grant for discovery of novel drug targets for treating MDR infections. Congrats!
- Anushree presents five talks on synthetic biology, antimicrobial adaptive resistance, and strategies for novel therapeutics for MDR bacterial infections at AIChE 2015 conference.
- Our paper featured in the Gram Negative Issue of ACS Infectious Disease!